
AMPfun is a web server prediction tool providing users an effective identification of antimicrobial peptides (AMPs) with their functional activities.
The antimicrobial peptides (AMPs) are produced by virtually all organisms on earth and are essential components of the innate immune system, which makes them the first line of defense against microbes in many organisms. Specifically, AMPs would disrupt the cell membrane of microbes or intracellular functions to kill them. The following figure is the schematic representation of some action mechanisms of membrane-active AMPs. Consequently, they are able to resist various pathogenic microorganisms, such as viruses, parasites, bacteria, and fungi. In general, AMPs consist of 10-50 amino acids and have little sequence homology to one another.
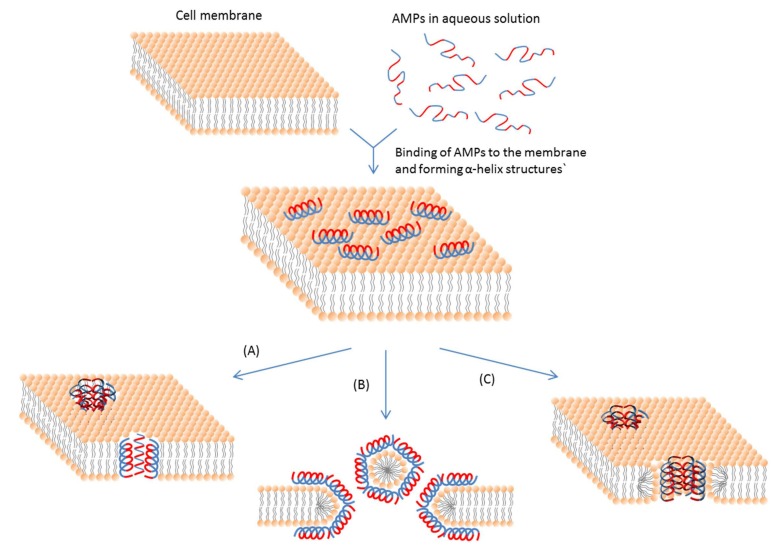
(A) Barrel-Stave model. AMP molecules insert themselves into the membrane perpendicularly. (B) Carpet model. Small areas of the membrane are coated with AMP molecules with hydrophobic sides facing inward leaving pores behind in the membrane. (C) Toroidal pore model. This model resembles the Barrel-stave model, but AMPs are always in contact with phospholipid head groups of the membrane. The blue color represents the hydrophobic portions of AMPs, while the red color represents the hydrophilic parts of the AMPs.
Ref. Bahar AA and Ren D (2013) Antimicrobial peptides.Pharmaceuticals (Basel). 2013 Nov 28;6(12):1543-75. doi: 10.3390/ph6121543.
The abuse of antibiotics has resulted in microbial pathogens developing resistance to chemical antibiotics and one result of this has been an urgent need to develop new therapeutics that can be used to treat infections. This seemingly intractable problem means that the remarkable benefits brought about by antibiotics is now seriously threatened by the rapid development of antibiotic resistant bacteria and this has developed into a serious global crisis. Compared to conventional antibiotics, AMPs have a basically simple chemical structure and this seem to mean that there is a lower probability of resistance emerging. As a result, AMPs have become attractive alternative when developing new therapeutics agents.
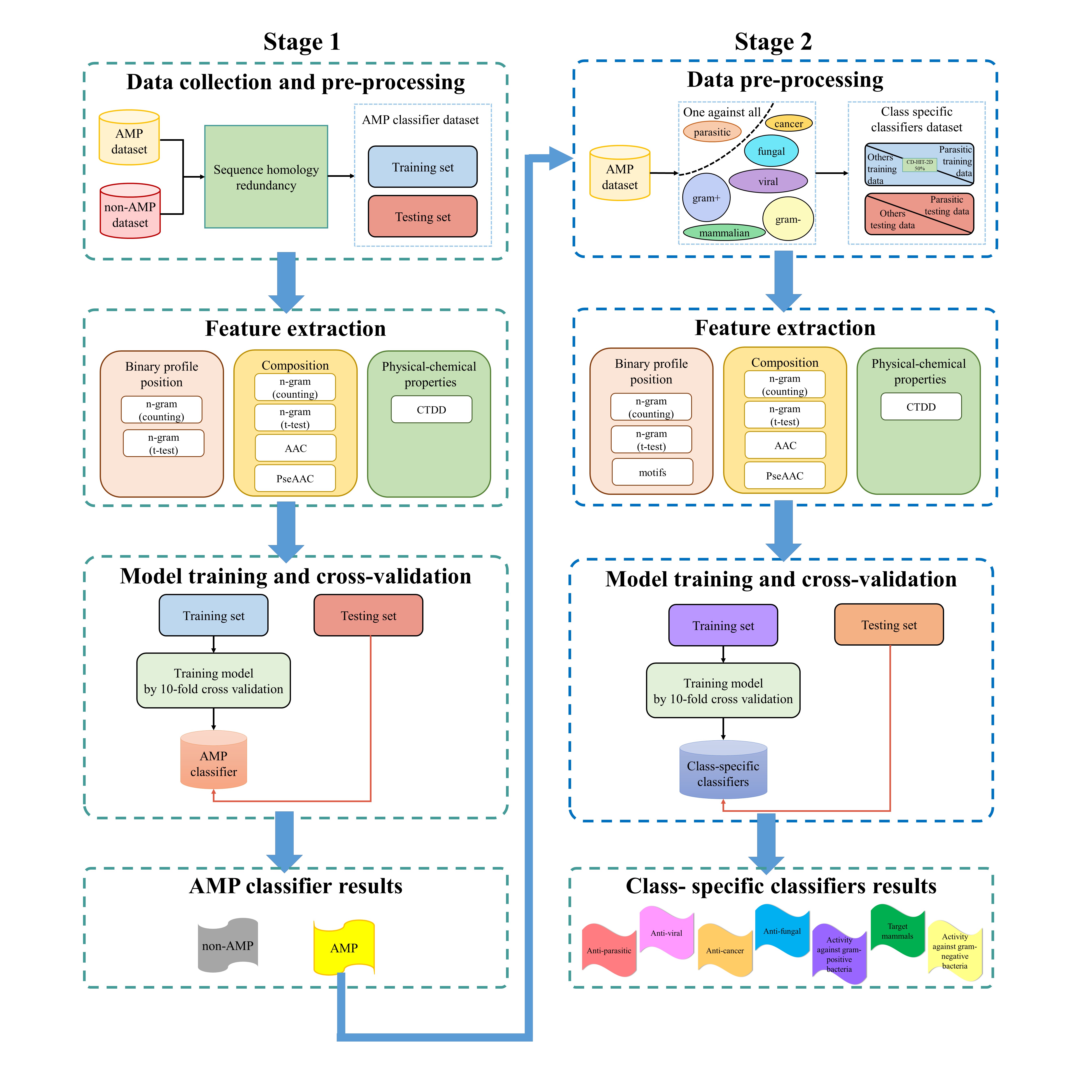
We adopted a two-stage framework based on machine learning technique that is able to identify AMPs and determine their functional activities. The flow chart for constructing the classifiers is shown below.
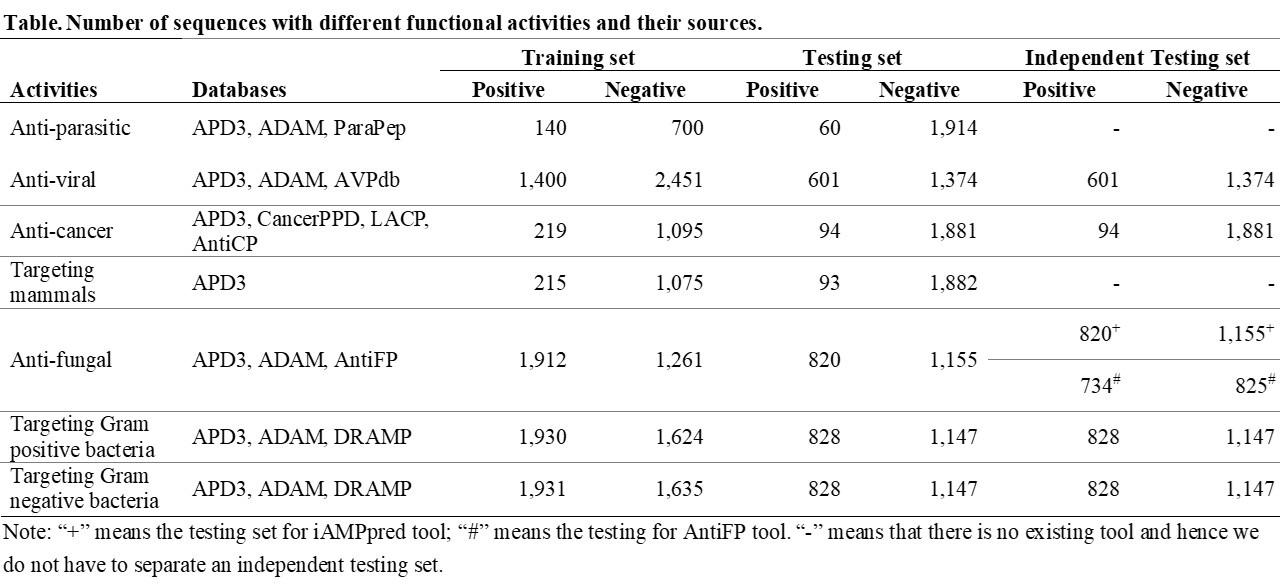
The following table demonstrates the total number of data that we used in this study.
It was extremely important to transform these sequences, which consisted of strings of amino acids, to reasonable and representative numerical values before building the prediction model. In this study, we divided the approaches into three types, namely binary profiling of amino acid position, amino acid composition, and physical-chemical properties. The binary profiling of amino acid position included n-gram binary profiling of position as determined by counting (NCB), n-gram binary profiling of position as determined by t-test (NTB), and motif based binary profiling of position (MB). The n-gram composition by counting (NCC), the n-gram composition by t-test (NTC), motifs composition (MC), and amino acid composition (AAC) are members of the composition category. Both pseudo amino acid composition (PseAAC) and composition transition distribution (CTDD) are physical-chemical properties. These aforementioned features and the detail descriptions of each of these features is presented below.
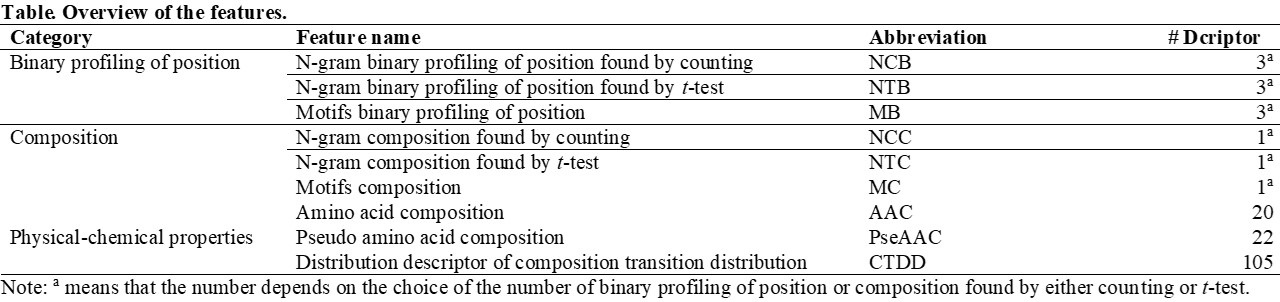
Furthermore, the forward feature selection algorithm is able to show that it is important to consider the crucial features when developing the classifiers on different AMP activities. To obtain better and more information from the peptides sequences in this study, we adopted the n-grams concept in order to encode the binary profiling of the positional and composition features. In addition, AAC, PseAAC, and CTDD are also used as features. Among the above three features, both AAC and PseAAC are commonly used features when separating AMPs and non-AMPs, and also when identifying their functional activity.